First, because the same recognition sequence occurs on both strands of the DNA duplex, the restriction endonuclease can bind to and cleave both strands, effectively cutting the DNA in half. This ability to cut across both strands is almost certainly the reason that restriction endonucleases have evolved to recognize nucleotide sequences with twofold rotational symmetry.
Second, because the bond cleaved by a restriction endonuclease is typically not positioned in the center of the recognition sequence to which it binds, and because the DNA strands are antiparallel, the cut sites for the two strands of a duplex are offset from each other (figure 2). After cleavage, each DNA fragment has a single-stranded end a few nucleotides long. The single-stranded ends of the two fragments are complementary to each other.
Why Restriction Endonucleases Are So Useful
There are hundreds of bacterial restriction endonucleases, and each one has a specific recognition sequence. By chance, a particular endonuclease’s recognition sequence is likely to occur somewhere in any given sample of DNA; the shorter the sequence, the more often it will arise by chance within a sample. Therefore, a given restriction endonuclease can probably cut DNA from any source into fragments. Each fragment will have complementary single stranded ends characteristic of that endonuclease. Because of their complementarity, these single-stranded ends can pair with each other (consequently, they are sometimes called “sticky ends”). Once their ends have paired, two fragments can then be joined together with the aid of the enzyme DNA ligase, which re-forms the phosphodiester bonds of DNA. What makes restriction endonucleases so valuable for genetic engineering is the fact that any two fragments produced by the same restriction endonuclease can be joined together. Fragments of elephant and ostrich DNA cleaved by the same endonuclease can be joined to one another as readily as two bacterial DNA fragments.
Genetic engineering involves manipulating specific genes by cutting and rearranging DNA. A restriction endonuclease cleaves DNA at a specific site, generating in most cases two fragments with short single-stranded ends. Because these ends are complementary to each other, any pair of fragments produced by the same endonuclease, from any DNA source, can be joined together.
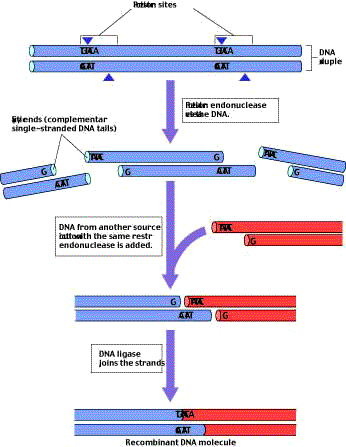 Figure 2
Figure 2
Many restriction endonucleases produce DNA fragments with “sticky ends.” The restriction endonuclease EcoRI always cleaves the sequence GAATTC between G and A. Because the same sequence occurs on both strands, both are cut. However, the two sequences run in opposite directions on the two strands. As a result, single-stranded tails are produced that are complementary to each other, or “sticky.”