Using Restriction Endonucleases to Manipulate Genes
A chimera is a mythical creature with the head of a lion, body of a goat, and tail of a serpent. Although no such creatures existed in nature, biologists have made chimeras of a more modest kind through genetic engineering.
Constructing pSC101
One of the first chimeras was manufactured from a bacterial plasmid called a resistance transfer factor by American geneticists Stanley Cohen and Herbert Boyer in 1973. Cohen and Boyer used a restriction endonuclease called EcoRI, which is obtained from Escherichia coli, to cut the plasmid into fragments. One fragment, 9000 nucleotides in length, contained both the origin of replication necessary for replicating the plasmid and a gene that conferred resistance to the antibiotic tetracycline (tetr). Because both ends of this fragment were cut by the same restriction endonuclease, they could be ligated to form a circle, a smaller plasmid Cohen dubbed pSC101 (figure 3).
Using pSC101 to Make Recombinant DNA
Cohen and Boyer also used EcoRI to cleave DNA that coded for rRNA that they had isolated from an adult amphibian, the African clawed frog, Xenopus laevis. They then mixed the fragments of Xenopus DNA with pSC101 plasmids that had been “reopened” by EcoRI and allowed bacterial cells to take up DNA from the mixture. Some of the bacterial cells immediately became resistant to tetracycline, indicating that they had incorporated the pSC101 plasmid with its antibiotic-resistance gene. Furthermore, some of these pSC101-containing bacteria also began to produce frog ribosomal RNA! Cohen and Boyer concluded that the frog rRNA gene must have been inserted into the pSC101 plasmids in those bacteria. In other words, the two ends of the pSC101
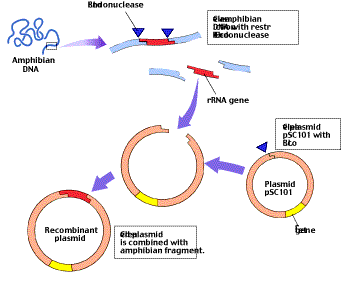 Figure 3
Figure 3
One of the first genetic engineering experiments. This diagram illustrates how Cohen and Boyer inserted an amphibian gene encoding rRNA into pSC101. The plasmid contains a single site cleaved by the restriction endonuclease EcoRI; it also contains tetr, a gene which confers resistance to the antibiotic tetracycline. The rRNA-encoding gene was inserted into pSC101 by cleaving the amphibian DNA and the plasmid with EcoRI and allowing the complementary sequences to pair.
plasmid, produced by cleavage with EcoRI, had joined to the two ends of a frog DNA fragment that contained the rRNA gene, also cleaved with EcoRI.
The pSC101 plasmid containing the frog rRNA gene is a true chimera, an entirely new genome that never existed in nature and never would have evolved by natural means. It is a form of recombinant DNA, that is, DNA created in the laboratory by joining together pieces of different genomes to form a novel combination.
Other Vectors
The introduction of foreign DNA fragments into host cells has become common in molecular genetics. The genome that carries the foreign DNA into the host cell is called a vector. Plasmids, with names like pUC18 can be induced to make hundreds of copies of themselves and thus of the foreign genes they contain. Much larger pieces of DNA can be introduced using YAKs (yeast artificial chromosomes) as a vector instead of a plasmid. Not all vectors have bacterial targets. Animal viruses such as the human cold virus adenovirus, for example, are serving as vectors to carry genes into monkey and human cells, and animal genes have even been introduced into plant cells.
One of the first recombinant genomes produced by genetic engineering was a bacterial plasmid into which an amphibian ribosomal RNA gene was inserted. Viruses can also be used as vectors to insert foreign DNA into host cells and create recombinant genomes.