What the Human Genome Is Like
Geography of the Genome
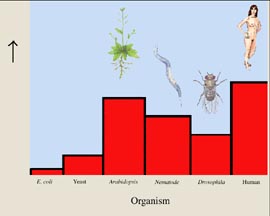
The number of genes in the human genome is only about 30,000 (figure 3). This is barely a third more than in nematodes, scarcely double the number in Drosophila, and but a quarter of the number that had been anticipated by scientists counting unique messenger RNA (mRNA) molecules.
How can human cells contain four times as many kinds of mRNA as there are genes? A typical human gene is not simply a straight sequence of DNA, with the order of its units corresponding to the sequence of amino acids in a protein. Instead, a human gene is fragmented. The sequence of DNA nucleotides that specifies a protein is broken into many bits called exons, scattered among much longer segments of nontranscribed DNA called introns. Imagine this paragraph was a human gene; all the occurrances of the letter e could be considered exons, while the rest would be noncoding introns.
When a cell uses a human gene to make a protein, it first manufactures mRNA copies of all the exons (protein- specifying fragments) of the gene, then splices the exons together. Now heres the turn-of-events researchers had not anticipated: the transcripts of human genes are often spliced together in different ways, called alternative splicing. Each exon is actually a module; one exon may code for one part of a protein, another for a different part of a protein. When the exon transcripts are mixed in different ways, very different protein shapes can be built.
With alternative mRNA splicing, it is easy to see how 30,000 genes can encode four times as many proteins. It seems that the added complexity of human proteins has been achieved not by gaining more gene parts, but rather by learning new ways to put them together. Great music is made from simple tunes in much the same way.
Genes are not distributed evenly over the genome. The small chromosome number 19 is packed densely with genes, transcription factors, and other functional elements. The much larger chromosomes numbers 4 and 8, by contrast, have few genes. On most chromosomes, vast stretches of seemingly barren DNA fill the chromosomes between scattered clusters rich in genes.
DNA That Codes for Proteins
Four different classes of protein-encoding genes are found in the human genome, differing largely in gene copy number.
Single-copy genes Many eukaryotic genes exist as single copies at a particular location on a chromosome. Mutations in these genes produce recessive Mendelian inheritance. Silent copies, inactivated by mutation, are called pseudogenes.
Figure 3 What the human genome is like.
The human genome has an unexpectedly small number of genes, some 30,000. This is not many more than the plant Arabidopsis, and only a third more than nematode worms.
Segmental duplications Sequencing of the human genome has revealed that human chromosomes contain many segmental duplications, where whole blocks of genes have been copied over from one chromosome to another. Blocks of similar genes in the same order are found throughout the genome, indicating segmental duplication has been a major factor in determining the human genomes present architecture. Chromosome 19 seems to have been the biggest borrower, with blocks of genes shared with 16 other chromosomes.
Multigene families As we have learned more about the nucleotide sequences of eukaryotic genomes, it has become apparent that many genes exist as parts of multigene families, groups of related but distinctly different genes that often occur together in a cluster. Multigene families differ from tandem clusters (see below) in that they contain far fewer genes (from three to several hundred), and those genes differ much more from one another than the genes in tandem clusters. Despite their differences, the genes in a multigene family are clearly related in their sequences, making it likely that they arose from a single ancestral sequence through a series of unequal crossing over events.
Tandem clusters A second class of repeated gene consists of DNA sequences that are repeated many times, one copy following another in tandem array. By transcribing all of the copies in these tandem clusters simultaneously, a cell can rapidly obtain large amounts of the product they encode. For example, the genes encoding rRNA are typically present in clusters of several hundred copies.