3. Biotechnology is producing a scientific revolution. DNA Sequence Technology
The 1980s saw an explosion of interest in biotechnology, the application of genetic engineering to practical human problems. Let us examine some of the major areas where these techniques have been put to use.
Genome Sequencing
Genetic engineering techniques are enabling us to learn a great deal more about the human genome. Several clonal libraries of the human genome have been assembled, using large-size restriction fragments. Any cloned gene can now be localized to a specific chromosomal site by using probes to detect in situ hybridization (that is, binding between the probe and a complementary sequence on the chromosome). Genes are now being mapped at an astonishing rate: genes that contribute to dyslexia, obesity, and cholesterol-proof blood are some of the important ones that were mapped in 1994 and 1995 alone! With an understanding of where specific genes are located in the human genome and how they work, it is not difficult to imagine a future in which virtually any genetic disease could be treated or perhaps even cured with gene therapy. As we mentioned in chapter 13, some success has already been reported in treating patients who have cystic fibrosis with a genetically corrected version of the cystic fibrosis gene.
An exciting scientific by-product of the human genome project has been the complete genome sequencing of many microorganisms with smaller genomes, on the order of a few Mb (table 1). In general, about half of the genes prove to have a known function; what the other half of the genes are doing is a complete mystery. The first eukaryotic genome to be sequenced in its entirety was that of brewer’s yeast Saccharomyces cerevisiae; many of its approximately 6000 genes have a similar structure to some human genes. The complete sequences of many much larger genomes have recently been completed, including the malarial Plasmodium parasite (30 Mb), the nematode (100 Mb), the plant Arabidopsis (100 Mb) (figure 13), the fruit fly Drosophila (120 Mb), and the mouse (300 Mb).
The international scientific community has over the last several years mounted a major effort to sequence the entire human genome. Because the human genome contains some 3000 Mb (million nucleotide base-pairs), this task has presented no small challenge. Rapid progress was made possible by the use of so-called shotgun cloning techniques, in which the entire genome is first fragmented, then each of the fragments is sequenced by automated machines, and finally computers use overlaps to order the fragments. All but a small portion of the sequence was completed by the beginning of the year 2000.
Table 1. Genome Sequencing Projects
Genome
Organism Size (Mb) Description
Archaebacteria
Methanococcus jannaschii 1.7 Extreme thermophile
Eubacteria
Escherichia coli 4.6 Laboratory standard
fungi
Saccharomyces cerevisiae 13 Baker’s yeast
protist
Plasmodium 30 Malarial parasite
plant
Arabidopsis thaliana 100 Relative of mustard plant
animal
Caenorhabditis elegans 100 Nematode
Drosophila melanogaster 120 Fruit fly
Mus musculus 300 Mouse
Homo sapiens 3,000 Human
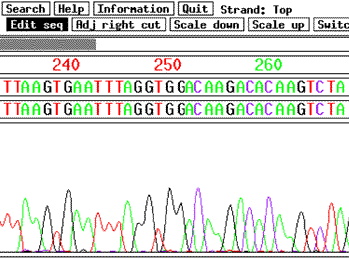 Figure 13
Figure 13
Part of the genome sequence of the plant Arabidopsis. Data from an automated DNA-sequencing run shows the nucleotide sequence for a small section of the Arabidopsis genome. Automated DNA sequencing has greatly increased the speed at which genomes can be sequenced.