Proteomics
Bioinformatics
Ideally, a researcher would like to be able to examine a nucleotide sequence and know what sort of functional protein the gene specifies. However, efforts to calculate what shape a protein will assume from knowledge of its amino acid sequence have proven difficult, even with the aid of large computers. However, by also looking at the proteins that are produced by the genes of the human genome, researchers are beginning to get a clearer picture of how gene sequence relates to protein shape and function.
Powerful computer programs are now having considerable success in screening the human genome for particular sorts of sequences, and increasing success in predicting the structure of a protein from the nucleotide sequence of the gene encoding it. This fast-growing area of genomics is loosely called bioinformatics. It combines molecular genetics and computational analysis in an attempt to predict what sort of protein a particular sequence encodes.
Proteins are three dimensional structures that often interact with other proteins and molecules to function. Thus, although the computer model provides a good starting point, a lot of protein biochemistry is still necessary to understand how the protein is actually working. For example, the structure of the Pax6 protein has been deduced from the genes nucleotide sequence, allowing researchers to predict how it might interact with one of the DNA sequences it regulates (figure 13). Determining the structures of all the genes Pax6 regulates can now be attempted using this approach.
Proteomics: The Next Frontier
With the sequencing of the human genome now essentially complete, researchers have begun an even more challenging task: the cataloging and analysis of every protein in the human body, an endeavor called proteomics. Each genes nucleotide sequence specifies an amino acid sequence that folds in a certain way, producing a protein whose shape gives it a particular function. Only by understanding the protein shapes that genes produce can we begin to make sense of the human genome.
Protein arrays, just like DNA microarrays, are now being developed to study all the proteins an organism possesses, its proteome. These arrays are screened using antibodies to specific proteins. Antibodies are fluorescently labeled so they can be detected, and the patterns on the protein array can then be determined by computer analysis. Technological advances are underway that will allow many proteins to be characterized on a mass scale in much less time than it took to uncover the structure of individual proteins like Pax6 in the past.
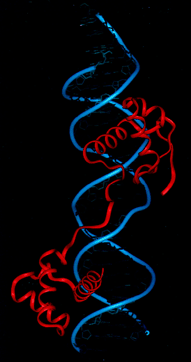
Figure 13 Pax6 protein interacting with DNA.
The red represents a ribbon running through the main carbon backbone of the Pax6 protein. The phosphates in the DNA backbone are colored blue. Two different domains in this protein allow it to bind to DNA where it initiates transcription.
Fortunately, while there may be as many as a million different proteins, most are just variations on a handful of themes. The same shared structural motifsbarrels, helices, molecular zippersare found in the proteins of plants, insects, and humans. The maximum number of distinct motifs has been estimated as fewer than 5000. About 1000 of these motifs have already been cataloged. Both public and privately-financed efforts are now underway to detail the shapes of all the common motifs.
Like, functional genomics, proteomics is a new approach that will enable analysis of proteins and comparisons at the protein level.