Identifying DNA: Southern Blotting
Once a gene has been cloned, it may be used as a probe to identify the same or a similar gene in another sample (figure 9). In this procedure, called a Southern blot, DNA from the sample is cleaved into restriction fragments with a restriction endonuclease, and the fragments are spread apart by gel electrophoresis. The double-stranded helix of each DNA fragment is then denatured into single strands by making the pH of the gel basic, and the gel is “blotted” with a sheet of nitrocellulose, transferring some of the DNA strands to the sheet. Next, a probe consisting of purified, single-stranded DNA corresponding to a specific gene (or mRNA transcribed from that gene) is poured over the sheet. Any fragment that has a nucleotide sequence complementary to the probe’s sequence will hybridize (base- pair) with the probe. If the probe has been labeled with 32P, it will be radioactive, and the sheet will show a band of radioactivity where the probe hybridized with the complementary fragment. 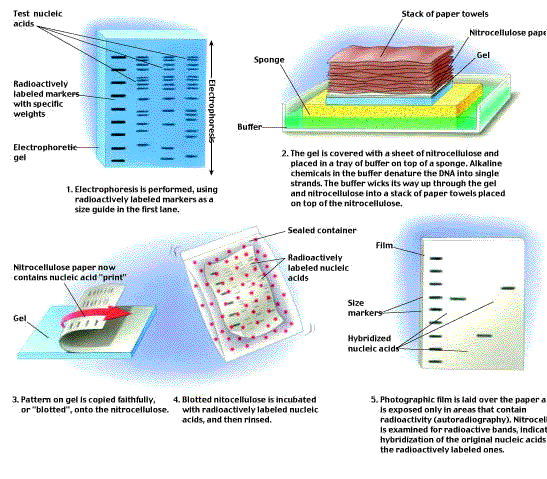 Figure 9
Figure 9
The Southern blot procedure. E. M. Southern developed this procedure in 1975 to enable DNA fragments of interest to be visualized in a complex sample containing many other fragments of similar size. The DNA is separated on a gel, then transferred (“blotted”) onto a solid support medium such as nitrocellulose paper or a nylon membrane. It is then incubated with a radioactive single-strand copy of the gene of interest, which hybridizes to the blot at the location(s) where there is a fragment with a complementary sequence. The positions of radioactive bands on the blot identify the fragments of interest.