Distinguishing Differences in DNA: RFLP Analysis
Often a researcher wishes not to find a specific gene, but rather to identify a particular individual using a specific gene as a marker. One powerful way to do this is to analyze restriction fragment length polymorphisms, or RFLPs (figure 10). Point mutations, sequence repetitions, and transposons that occur within or between the restriction endonuclease recognition sites will alter the length of the DNA fragments (restriction fragments) the restriction endonucleases produce. DNA from different individuals rarely has exactly the same array of restriction sites and distances between sites, so the population is said to be polymorphic (having many forms) for their restriction fragment patterns. By cutting a DNA sample with a particular restriction endonuclease, separating the fragments according to length on an electrophoretic gel, and then using a radioactive probe to identify the fragments on the gel, one can obtain a pattern of bands often unique for each region of DNA analyzed. These “DNA fingerprints” are used in forensic analysis during criminal investigations. RFLPs are also useful as markers to identify particular groups of people at risk for some genetic disorders.
Making an Intron-Free Copy of a Eukaryotic Gene
Eukaryotic genes are encoded in exons separated by numerous nontranslated introns. When the gene is transcribed to produce the primary transcript, the introns are cut out during RNA processing to produce the mature mRNA transcript. When transferring eukaryotic genes into bacteria, it is desirable to transfer DNA already processed this way, instead of the raw eukaryotic DNA, because bacteria lack the enzymes to carry out the processing. To do this, genetic engineers first isolate from the cytoplasm the mature mRNA corresponding to a particular gene. They then use an enzyme called reverse transcriptase to make a DNA version of the mature mRNA transcript (figure 11). The single strand of DNA can then serve as a template for the synthesis of a complementary strand. In this way, one can produce a double-stranded molecule of DNA that contains a gene lacking introns. This molecule is called complementary DNA, or cDNA. 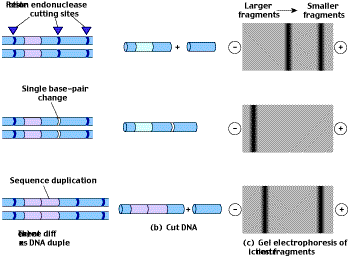 Figure 10
Figure 10
Restriction fragment length polymorphism (RFLP) analysis. (a) Three samples of DNA differ in their restriction sites due to a single base-pair substitution in one case and a sequence duplication in another case. (b) When the samples are cut with a restriction endonuclease, different numbers and sizes of fragments are produced. (c) Gel electrophoresis separates the fragments, and different banding patterns result.
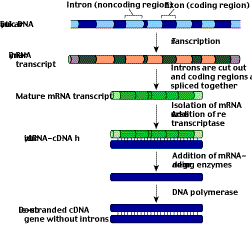 Figure 11
Figure 11
The formation of cDNA. A mature mRNA transcript is isolated from the cytoplasm of a cell. The enzyme reverse transcriptase is then used to make a DNA strand complementary to the processed mRNA. That newly made strand of DNA is the template for the enzyme DNA polymerase, which assembles a complementary DNA strand along it, producing cDNA, a double-stranded DNA version of the intron-free mRNA.